
An alternative method has been developed in which pronuclei are transplanted just after meiosis has been completed prior to the first mitotic division. This has been demonstrated in proof of concept studies, which have used an approach of transplanting pronuclei soon before the first mitotic division occurred however, it has been found that normally fertilised zygotes are unable to satisfactorily tolerate this approach ( Hyslop et al., 2016). Correctly diagnosing severe pathogenic mutations in mtDNA is also necessary when considering techniques such as in vitro fertilisation to prevent such mutations being passed on to future generations ( Gorman et al., 2015). Identifying pathogenic mutations in mitochondrial DNA (mtDNA) may also provide more effective diagnostic tools in addition to aiding disease prevalence studies, which can be used to inform healthcare budgets and public health strategies ( Gorman et al., 2015).
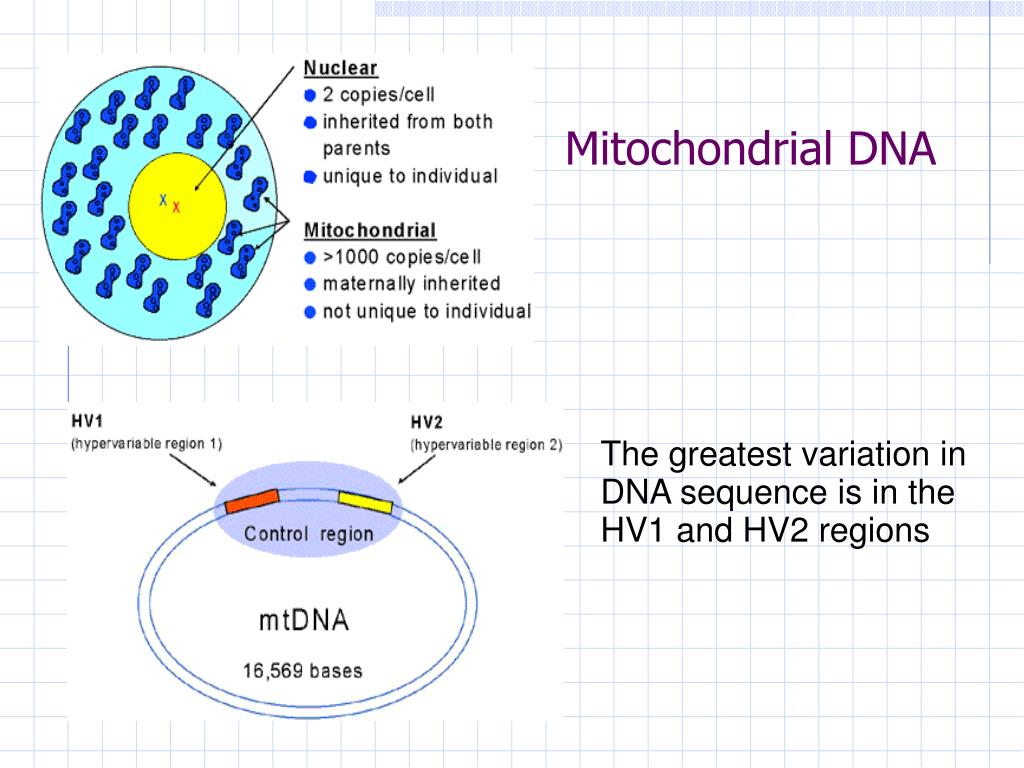
Developing and implementing effective treatments and preventative strategies for mitochondrial disease may improve the health of a substantial portion of the population ( Gorman et al., 2015). Mitochondrial diseases affect more than 10 per 100,000 individuals (including mitochondrial disorders caused by mutations in either the mitochondrial or nuclear genomes) ( Gorman et al., 2015).

This review extends a mini-review previous published by Bris et al. This may provide perspective for researchers seeking to develop new bioinformatics tools by informing them of the current gaps in the market, and therefore may provide inspiration for the development of new tools which would enable researchers to investigate the mitochondrial genome further. We also discuss challenges relating to the field of mitochondrial genomics and discuss bioinformatics capabilities which are currently lacking. We discuss and compare the capabilities offered by a variety of bioinformatics software ( Figures 1, 2), helping readers to make an informed choice as to which tools would best suit their needs. This brief overview highlights the importance of the mitochondrial genome from a scientific and clinical perspective, and also provides readers with signposts to the bioinformatics tools currently available. In this review, we discuss the role of the mitochondria in relation to disease, with a focus on mitochondrial genomics, and review the bioinformatic tools currently available for the analysis of the mitochondrial genome. Genetics resources are well established for phylogeny and DNA sequence changes, but further epigenetic and gene expression resources need to be developed for mitochondrial genomics. No single tool will provide all users with the resources they require therefore, the most suitable tool will vary between users depending on the nature of the work they aim to carry out. These resources fulfil a variety of functions, with some being highly specialised.
This ‘mitochondrial toolbox’ summary resource will enable researchers to rapidly identify the resource(s) most suitable for their needs. Twenty-four online resources dedicated to mitochondrial genomics were reviewed.
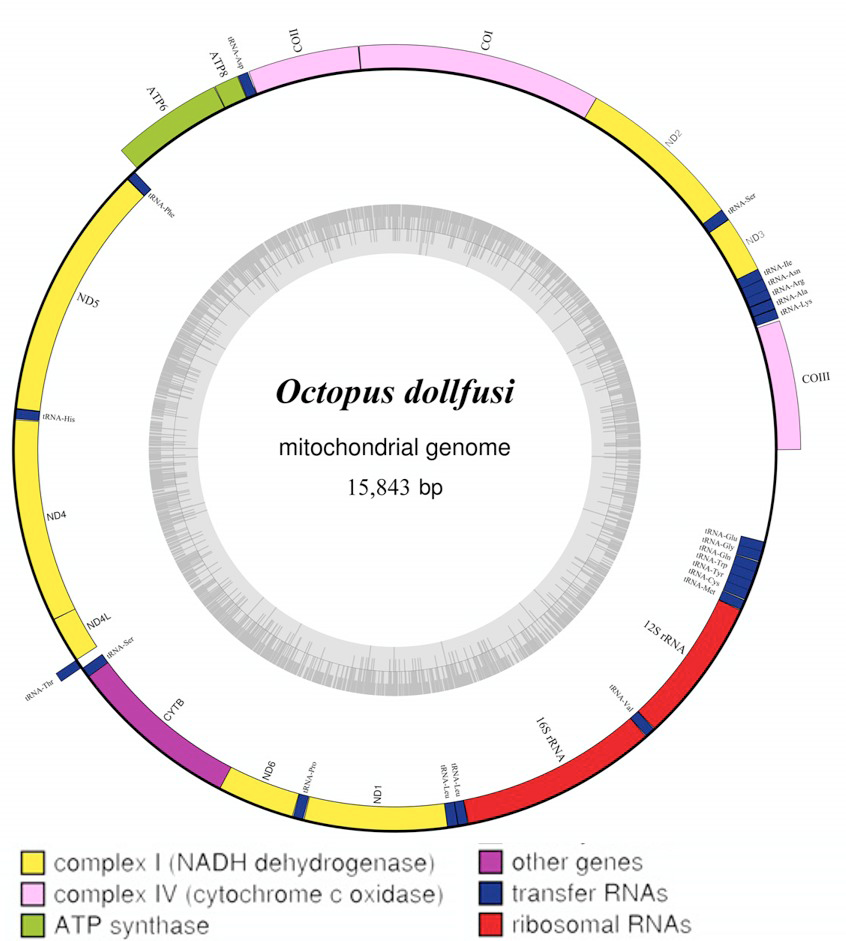
There is often a large amount of complex data generated (for example via next generation sequencing), which requires optimised bioinformatics tools to efficiently and effectively generate robust outcomes from these large datasets. There is emerging evidence that differences in the mitochondrial genome may contribute to multiple common diseases, leading to an increasing number of studies exploring mitochondrial genomics. Mitochondria play a significant role in many biological systems. 2School of Electronics, Electrical Engineering and Computer Science, Queen’s University Belfast, Belfast, United Kingdom.1Centre for Public Health, Institute of Clinical Sciences B, Queen’s University Belfast, Royal Victoria Hospital, Belfast, United Kingdom.Ruaidhri Cappa 1*, Cassio de Campos 2, Alexander P.


 0 kommentar(er)
0 kommentar(er)
